This notebook was created by Sergey Tomin (sergey.tomin@desy.de). April 2025.
Optics Design for High Time Resolution Measurements with TDS
This tutorial is motivated by a practical task: improving the time resolution of current profile measurements using a Transverse Deflecting Structure (TDS) at the European XFEL (EuXFEL).
The tutorial itself is available in Jupyter Notebook format and can be downloaded here.
The lattice files used in this tutorial can be found in this repository.
A Bit of Simple Theory
The transverse position of a particle along a beamline is given by:
where:
- is the betatron function at position ,
- is the betatron phase,
- .
Taking the derivative:
with .
At the TDS Position
Let’s assume the TDS is located at :
- The particle receives a transverse kick: ,
- The transverse position at the TDS is zero: .
Then:
From this, we get:
At the Screen
The transverse position on the screen becomes:
With and using the identity :
Transverse Kick from the Deflecting Structure
The kick from the TDS depends on time:
Assuming (zero-crossing), the rms beam size on the screen is:
Time Resolution of the TDS
Streaking (Calibration) Factor
The streaking factor is:
Note that this expression can be written more compactly by recognizing that the corresponding element of the transfer matrix is:
or, if the streaking occurs in the vertical direction:
Thus, the streaking factor simplifies to:
Time Resolution
The time resolution is defined as:
Using , we get:
So the time resolution depends only on:
- emittance
- voltage
- wavelength
- beta function at the TDS
- phase advance between TDS and screen
Practical Example: Optimizing Time Resolution with TDS at EuXFEL
In this section, we apply the theory from the previous part to a real EuXFEL lattice using Ocelot.
import sys
sys.path.append("/Users/tomins/Nextcloud/DESY/repository/ocelot/")
import os
import copy
import pandas as pd
from ocelot import *
from ocelot.gui import *
import l2, l3 # lattices can be found in https://github.com/ocelot-collab/EuXFEL-Lattice/tree/main/lattices/longlist_2024_07_04
initializing ocelot...
Check design optics
lat_l2 = MagneticLattice(l2.cell + l3.cell, stop=l3.bpmc_488_l3) # id_32072837_ - Drift in front of first A6 RF module
tws = twiss(lat_l2, tws0=l2.tws0)
plot_opt_func(lat_l2, tws, top_plot=["Dy"], legend=False)
plt.savefig("L2_design.png")
plt.show()
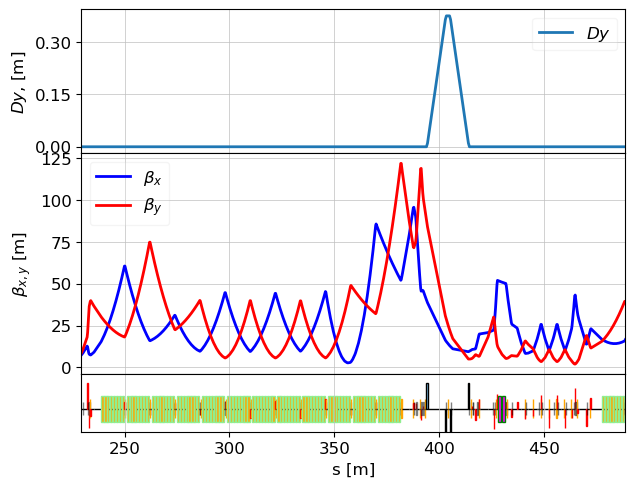
Check Twiss Parameters at Key Elements
We use markers for the TDS and screens (e.g., marker_tds_b2, otrb_457_b2) and inspect relevant optics values like beta functions and phase advances.
tws = twiss(lat_l2, tws0=l2.tws0, attach2elem=True)
# with attach2elem=True to all elements will be attached Twiss object in element.tws
# let's print beta_x
print(l2.ensub_466_b2.tws.beta_y)
5.058664837892
Define Matching Start and End Points
We preserve Twiss parameters at match_385_b2 (entry point after L2) and id_32072837_ (end of lattice).
END_ELEM = l3.id_32072837_
tws_match_385 = copy.deepcopy(l2.match_385_b2.tws)
tws_end = copy.deepcopy(END_ELEM.tws)
Shorten Lattice to Relevant Region
We exclude upstream quadrupoles and start optimization just after L2.
lat = MagneticLattice(l2.cell+l3.cell, start=l2.match_385_b2, stop=END_ELEM)
tws_des = twiss(lat, tws0=tws_match_385)
plot_opt_func(lat, tws_des, top_plot = ["Dy"], legend=False)
plt.savefig("TDS_area_design.png")
plt.show()
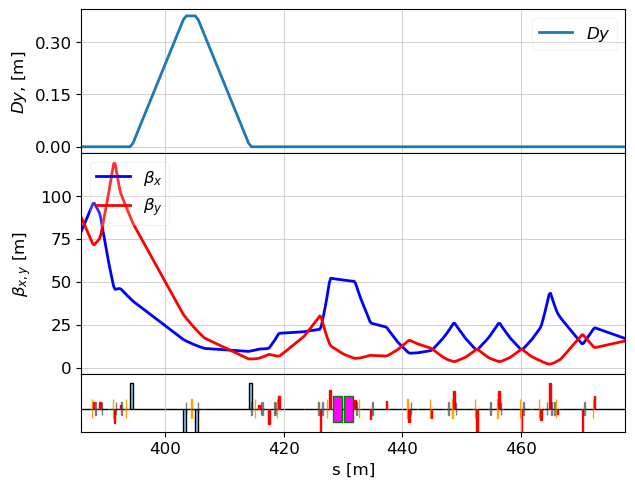
(Optional) Save Quadrupole Strengths for Reference
We optionally store the design quadrupole strengths in a CSV file for comparison later. The function looks a bit complicated just because I wanted to avoid overwriting every time design quads strengths.
# let's save design kicks to a dictionary
df_filename = "quads_strengths.csv"
design_column = "design"
if os.path.exists(df_filename):
quads_kicks_df = pd.read_csv(df_filename, index_col=0)
if design_column in quads_kicks_df.columns:
print(f"Column '{design_column}' already exists. Skipping step.")
else:
print(f"Column '{design_column}' not found. Proceeding to add it.")
quads_kicks_df[design_column] = pd.Series(d_design)
df.to_csv(df_filename)
else:
print("File does not exist. Creating new DataFrame.")
# let's save design kicks to a dictionary
d_design = {}
for e in lat.sequence:
if e.__class__ == Quadrupole:
d_design[e.id] = e.k1
quads_kicks_df = pd.DataFrame({design_column: d_design})
quads_kicks_df.to_csv(df_filename, index=True)
Column 'design' already exists. Skipping step.
Display Twiss Parameters at Specific Elements
We define a helper function to show selected optics values and compute R12 matrix elements.
It can be done in different ways but we will use pandas.
# List of elements where we want to see Twiss parameters
elements_for_comparision = {'TDS 429': l2.marker_tds_b2, "Scr 450": l2.otrb_450_b2, "Scr 454": l2.otrb_454_b2, 'Scr 457': l2.otrb_457_b2, 'Scr 461': l2.otrb_461_b2, 'end': END_ELEM}
# Attributes we want to compare
attributes = ['beta_x', 'beta_y', 'alpha_x', 'alpha_y', 'mux', "muy"]
def table_update(lat, tws0, elements_for_comparision, attributes):
# calculate Twiss
tws = twiss(lat, tws0=tws0, attach2elem=True)
# Build the table from tws list
table = pd.DataFrame({name: [getattr(getattr(obj, "tws"), attr) for attr in attributes] for name, obj in elements_for_comparision.items()},
index=attributes)
# make phase advance in degree
table.loc['mux'] = (table.loc['mux'] - table.loc['mux', 'TDS 429'])*180/np.pi
table.loc['muy'] = (table.loc['muy'] - table.loc['muy', 'TDS 429'])*180/np.pi
# add R12 elements into table
R12_values = copy.copy(elements_for_comparision)
for key in R12_values:
stop_elem = elements_for_comparision[key]
_, R, _ = lat.transfer_maps(energy=2.4, start=l2.marker_tds_b2, stop=stop_elem)
R12_values[key] = R[0, 1]
table.loc['R12'] = R12_values
return table
table = table_update(lat, tws_match_385, elements_for_comparision, attributes)
table
| TDS 429 | Scr 450 | Scr 454 | Scr 457 | Scr 461 | end | |
|---|---|---|---|---|---|---|
| beta_x | 50.97858 | 17.07336 | 16.94874 | 17.04623 | 17.18839 | 16.88229 |
| beta_y | 7.97626 | 5.96935 | 6.03083 | 5.97761 | 5.98449 | 15.53650 |
| alpha_x | 0.22863 | 2.13837 | -2.15339 | 2.13443 | -2.18046 | 0.43030 |
| alpha_y | 0.89572 | -0.97954 | 1.00367 | -0.98736 | 0.98641 | -0.60920 |
| mux | 0.00000 | 71.68294 | 88.38849 | 98.69572 | 115.29531 | 157.32295 |
| muy | 0.00000 | 166.8764 | 193.9847 | 243.5603 | 270.7594 | 407.94331 |
| R12 | 0.00000 | 28.00731 | 29.38263 | 29.13983 | 26.76309 | 11.31032 |
Matching
Objective: High Beta at TDS and 90° Phase Advance to Screen
We now want to modify the optics such that:
- The beta function at the TDS position is large (e.g., 120 m), which improves time resolution.
- The phase advance between the TDS and screen is exactly 90 degrees.
To achieve this, we define a set of constraints and a list of quadrupoles we allow the matcher to modify.
constr = {
l2.marker_tds_b2: {"beta_x": 150, "alpha_x": 0},
l2.otrb_457_b2: {"beta_x": 17},
"delta": {
l2.marker_tds_b2: ["mux", 0],
l2.otrb_457_b2: ["mux", 0],
"val": np.pi / 2,
"weight": 1_000_007
},
}
vars = [
l2.qd_417_b2,
l2.qd_418_b2, l2.qd_425_b2, l2.qd_427_b2,
l2.qd_431_b2, l2.qd_434_b2, l2.qd_437_b2, l2.qd_440_b2,
l2.qd_444_b2, l2.qd_448_b2, l2.qd_452_b2, l2.qd_456_b2
]
match(lat, constr, vars, tw=tws_match_385, verbose=False, max_iter=1000, method='simplex')
tws = twiss(lat, tws0=tws_match_385)
plot_opt_func(lat, tws, top_plot=["Dy"], legend=False)
plt.show()
initial value: x = [-0.7502347655006337, 0.6491929171989861, -1.3008034, 0.9414835846007605, 0.43518302749894383, -0.5278581910012674, 0.4055492834980989, -0.6685246719983101, -0.4582186614997888, 0.8960955489987327, -1.263284384000845, 0.8960955489987327] Optimization terminated successfully. Current function value: 0.000031 Iterations: 566 Function evaluations: 892
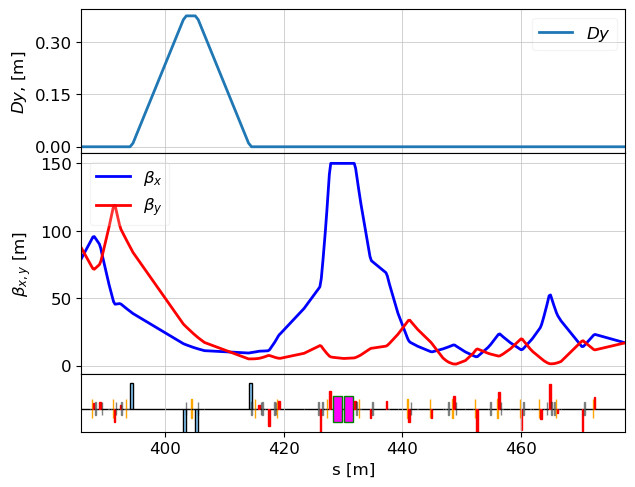
table = table_update(lat, tws_match_385, elements_for_comparision, attributes)
table
| TDS 429 | Scr 450 | Scr 454 | Scr 457 | Scr 461 | end | |
|---|---|---|---|---|---|---|
| beta_x | 149.99999 | 10.17169 | 13.46432 | 17.00000 | 20.59933 | 16.91344 |
| beta_y | 5.39319 | 3.80610 | 9.25716 | 12.53528 | 10.35206 | 16.98702 |
| alpha_x | 0.00001 | 1.20565 | -2.32524 | 1.64110 | -2.78659 | 0.43173 |
| alpha_y | 0.07739 | -1.48590 | 0.75004 | -1.76435 | 2.13902 | -0.74985 |
| mux | 0.00000 | 53.13755 | 78.53762 | 90.00020 | 104.86418 | 144.32068 |
| muy | 0.00000 | 189.1443 | 215.2306 | 240.5480 | 254.87936 | 415.89824 |
| R12 | 0.00000 | 31.25176 | 44.04417 | 50.49753 | 53.72675 | 29.37750 |
Matching to Final Conditions
We now restore the beam optics to match the original design values at the end of the beamline.
constr_end = {
END_ELEM: {
"beta_x": tws_end.beta_x,
"beta_y": tws_end.beta_y,
"alpha_x": tws_end.alpha_x,
"alpha_y": tws_end.alpha_y
},
# "delta": {
# l2.otrb_457_b2: ["mux", 0],
# END_ELEM: ["mux", 0],
# "val": (790-690.22)/180*np.pi,
# "weight": 1_000_007
# },
}
vars_end = [
l2.qd_459_b2,
l2.qd_463_b2, l2.qd_464_b2, l2.qd_465_b2,
l3.qd_470_b2, l3.qd_472_b2
]
match(lat, constr_end, vars_end, tw=tws_match_385, verbose=False, max_iter=2000, method='simplex')
tws_hi_res = twiss(lat, tws0=tws_match_385)
plot_opt_func(lat, tws_hi_res, top_plot=["Dy"], legend=False)
plt.show()
table = table_update(lat, tws_match_385, elements_for_comparision, attributes)
table
initial value: x = [-1.263284384000845, -0.569607097600338, 1.2982678500000002, -0.24686100550063372, -1.1289067389987326, 0.6611797761005492] Optimization terminated successfully. Current function value: 0.000049 Iterations: 870 Function evaluations: 1359
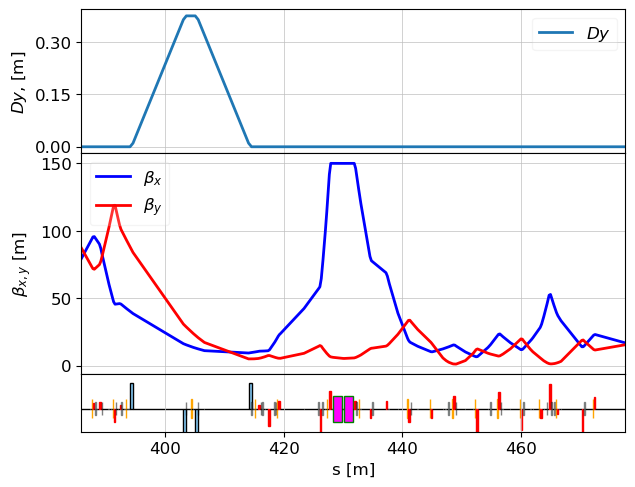
| TDS 429 | Scr 450 | Scr 454 | Scr 457 | Scr 461 | end | |
|---|---|---|---|---|---|---|
| beta_x | 149.99999 | 10.17169 | 13.46432 | 17.00000 | 20.71699 | 16.88229 |
| beta_y | 5.39319 | 3.80610 | 9.25716 | 12.53528 | 10.24631 | 15.53648 |
| alpha_x | 0.00001 | 1.20565 | -2.32524 | 1.64110 | -2.82521 | 0.43030 |
| alpha_y | 0.07739 | -1.48590 | 0.75004 | -1.76435 | 2.15514 | -0.60920 |
| mux | 0.00000 | 53.13755 | 78.53762 | 90.00020 | 104.84392 | 144.39113 |
| muy | 0.00000 | 189.1443 | 215.2306 | 240.5480 | 254.9183 | 417.5666 |
| R12 | 0.00000 | 31.25176 | 44.04417 | 50.49753 | 53.88501 | 29.30015 |
Compare design and new optics
bx_n = [tw.beta_x for tw in tws_hi_res]
by_n = [tw.beta_y for tw in tws_hi_res]
s_n = np.array([tw.s for tw in tws_hi_res])
bx_d = [tw.beta_x for tw in tws_des]
by_d = [tw.beta_y for tw in tws_des]
s_d = np.array([tw.s for tw in tws_des])
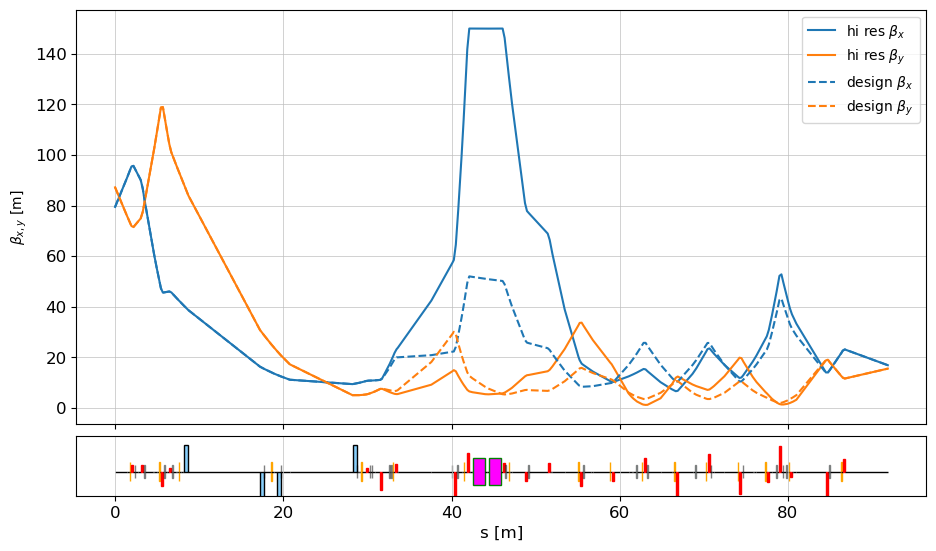
fig, ax = plot_API(lat, legend=False, figsize=[10,6])
ax.plot(s_n - s_n[0], bx_n, 'C0', label=r"hi res $\beta_{x}$ ")
ax.plot(s_n - s_n[0], by_n, 'C1', label=r"hi res $\beta_{y}$ ")
ax.plot(s_d - s_d[0], bx_d, "C0--", label=r"design $\beta_{x}$ ")
ax.plot(s_d - s_d[0], by_d, "C1--", label=r"design $\beta_{y}$ ")
ax.set_ylabel(r"$\beta_{x,y}$ [m]")
ax.legend()
#plt.savefig("TDS_90m.png")
plt.show()
for key in ['beta_x', "beta_y", "alpha_x", "alpha_y"]:
print(key, " :", getattr(tws_hi_res[-1], key), getattr(tws_des[-1], key))
beta_x : 16.882291323377554 16.882288192819743 beta_y : 15.536480497394853 15.536499055076685 alpha_x : 0.4302991727930746 0.4303020309015204 alpha_y : -0.6092012060842842 -0.609204409231985
Write to dataframe new quads kicks. Change name of new_colimn
WRITE_TO_FILE = True
REWRITE = True
beta_tds_ampl = constr[l2.marker_tds_b2]["beta_x"]
new_column = f'TDS {beta_tds_ampl}m'
quads_kicks_df = pd.read_csv(df_filename, index_col=0)
# let's save design kicks to a dictionary
d_new = {}
for e in lat.sequence:
if e.__class__ == Quadrupole:
d_new[e.id] = e.k1
if WRITE_TO_FILE:
if new_column in quads_kicks_df.columns and not REWRITE:
print(f"Column '{new_column}' already exists. Skipping step.")
else:
print(f"Column '{new_column}' not found. Proceeding to add it.")
quads_kicks_df[new_column] = pd.Series(d_new)
quads_kicks_df.to_csv(df_filename)
quads_kicks_df
Column 'TDS 140m' already exists. Skipping step.
| Quad | design | TDS 70m | TDS 90m | TDS 120m | TDS 140m | TDS 150m |
|---|---|---|---|---|---|---|
| QD.387.B2 | 0.335173 | 0.335173 | 0.335173 | 0.335173 | 0.335173 | 0.335173 |
| QD.388.B2 | 0.355996 | 0.355996 | 0.355996 | 0.355996 | 0.355996 | 0.355996 |
| QD.391.B2 | -0.725525 | -0.725525 | -0.725525 | -0.725525 | -0.725525 | -0.725525 |
| QD.392.B2 | 0.196996 | 0.196996 | 0.196996 | 0.196996 | 0.196996 | 0.196996 |
| QD.415.B2 | 0.180686 | 0.180686 | 0.180686 | 0.180686 | 0.180686 | 0.180686 |
| QD.417.B2 | -0.750235 | -0.784398 | -0.844188 | -0.925253 | -0.878182 | -0.971459 |
| QD.418.B2 | 0.649193 | 0.584715 | 0.529610 | 0.462214 | 0.337036 | 0.409773 |
| QD.425.B2 | -1.300803 | -1.347249 | -1.304467 | -1.251303 | -1.156159 | -1.308911 |
| QD.427.B2 | 0.941484 | 0.961247 | 0.959889 | 0.955579 | 0.929976 | 0.990706 |
| QD.431.B2 | 0.435183 | 0.438339 | 0.448529 | 0.446643 | 0.500821 | 0.443519 |
| QD.434.B2 | -0.527858 | -0.516594 | -0.534249 | -0.520934 | -0.612066 | -0.505477 |
| QD.437.B2 | 0.405549 | 0.424919 | 0.435292 | 0.444177 | 0.424977 | 0.443716 |
| QD.440.B2 | -0.668525 | -0.694907 | -0.699210 | -0.737736 | -0.726803 | -0.745826 |
| QD.444.B2 | -0.458219 | -0.453644 | -0.463892 | -0.485140 | -0.462831 | -0.499049 |
| QD.448.B2 | 0.896096 | 0.830044 | 0.788204 | 0.758349 | 0.714462 | 0.711105 |
| QD.452.B2 | -1.263284 | -1.280195 | -1.307186 | -1.370400 | -1.379105 | -1.412015 |
| QD.456.B2 | 0.896096 | 0.914291 | 0.919221 | 0.908725 | 0.987346 | 0.937405 |
| QD.459.B2 | -1.263284 | -1.365812 | -1.289895 | -1.160471 | -1.135981 | -1.168825 |
| QD.463.B2 | -0.569607 | -0.516755 | -0.521682 | -0.541629 | -0.480187 | -0.536436 |
| QD.464.B2 | 1.298268 | 1.308125 | 1.316963 | 1.335647 | 1.230522 | 1.338992 |
| QD.465.B2 | -0.246861 | -0.235193 | -0.244567 | -0.263073 | -0.241534 | -0.264049 |
| QD.470.B2 | -1.128907 | -1.199967 | -1.210101 | -1.231144 | -1.292447 | -1.261294 |
| QD.472.B2 | 0.661180 | 0.650804 | 0.647835 | 0.650371 | 0.757629 | 0.660549 |
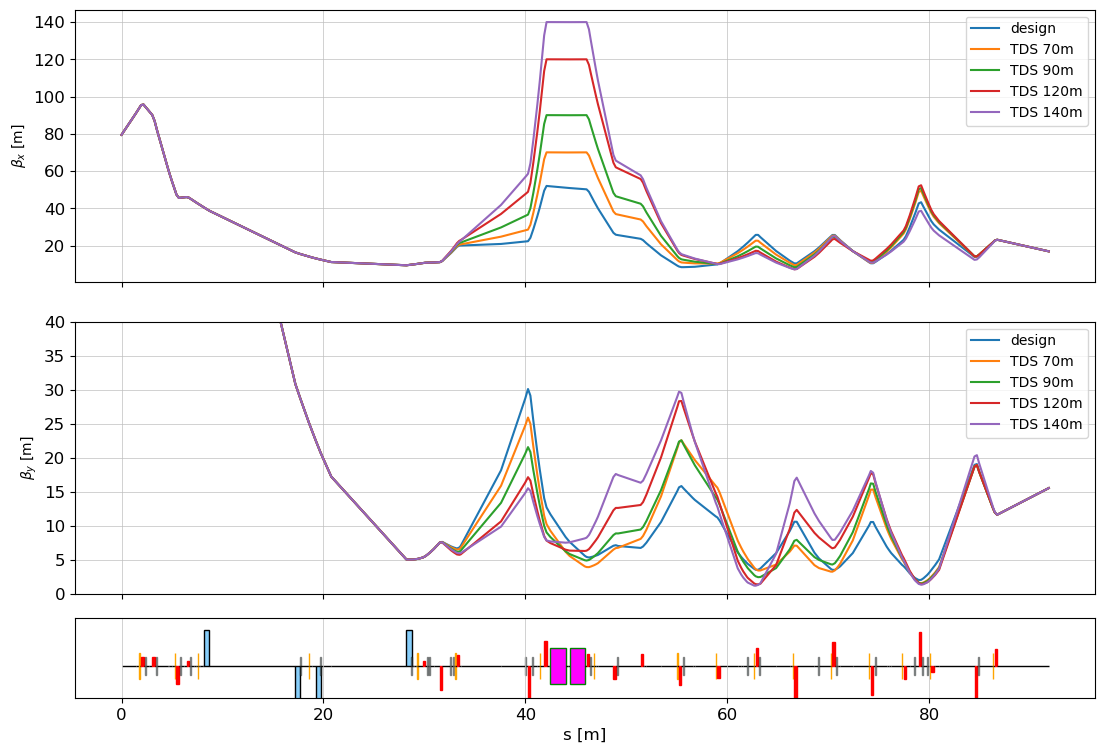
Check optics again from dataframe
quads_kicks_df = pd.read_csv(df_filename, index_col=0)
quads = list(quads_kicks_df.index)
optics = list(quads_kicks_df.columns)
fig, (ax_extra, ax_xy) = plot_API(lat, figsize=(12,8), add_extra_subplot=True)
ax_extra.set_ylabel(r"$\beta_x$ [m]")
ax_xy.set_ylabel(r"$\beta_y$ [m]")
data = {}
for opt in optics:
data[opt] = []
for e in lat.sequence:
if e.id in quads:
e.k1 = quads_kicks_df[opt][e.id]
tws = twiss(lat, tws0=tws_match_385)
data[opt] = tws
s = np.array([tw.s for tw in tws]) - tws[0].s
bx = [tw.beta_x for tw in tws]
by = [tw.beta_y for tw in tws]
ax_extra.plot(s, bx, label=opt)
ax_xy.plot(s, by, label=opt)
ax_xy.legend()
ax_extra.legend()
plt.show()
Let’s Put Some Numbers to Understand the TDS Voltage (Streaking/Calibration Factor)
The streaking factor is given by:
or, as defined above, it can be rewritten using the transfer matrix element:
During experimental study of the proposed optics, we measured a calibration factor of mm/ps for our S-band TDS (operating at 3 GHz). For optics TDS150m, we have between the TDS and a screen (Scr 457). Let’s calculate the required TDS voltage:
f = 3e9 # [Hz] frequency of the S-band TDS
S = 11.1e-3/1e-12 # [mm/ps] → [m/s]
R12 = 50.5
Lrf = speed_of_light /f
pc = 2400 # [MeV]
V = S * Lrf * pc /(R12 * 2 * np.pi* speed_of_light)
print(f"TDS voltage = {V} MV")
TDS voltage = 27.986057319921404 MV